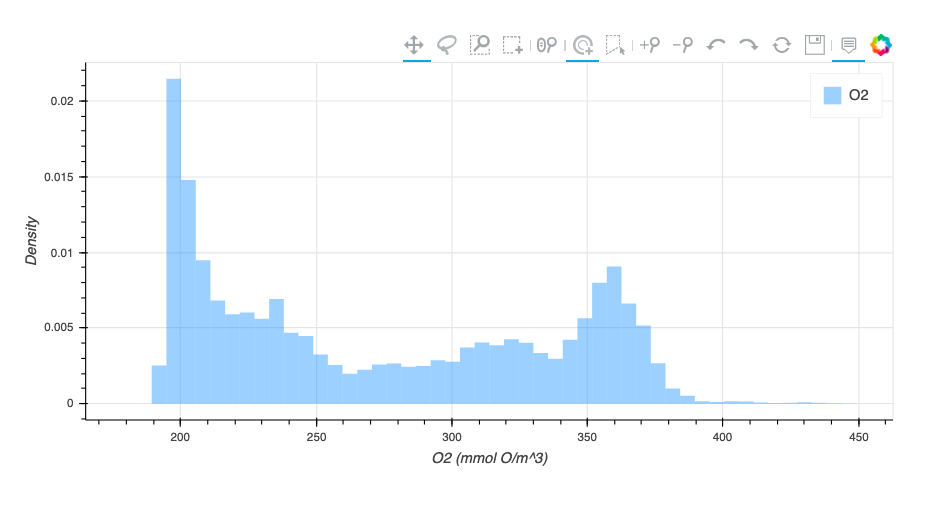
Histogram Method
Reformat netCDF4 File for a Function Call
NetCDF4-Compatible Function
Testing Function
xFile = xr.open_dataset('http://3.88.71.225:80/thredds/dodsC/las/id-a1d60eba44/data_usr_local_tomcat_content_cbiomes_20190510_20_darwin_v0.2_cs510_darwin_v0.2_cs510_nutrients.nc.jnl')
tables = [xFile] # see catalog.csv for the complete list of tables and variable names
variables = ['O2'] # see catalog.csv for the complete list of tables and variable names
startDate = '2010-04-30'
endDate = '2010-04-30'
lat1, lat2 = -80, 80
lon1, lon2 = -80, 80
depth1, depth2 = 0, 10
fname = 'DEP'
exportDataFlag = False # True if you you want to download data
xarrayPlotDist(tables, variables, startDate, endDate, lat1, lat2, lon1, lon2, depth1, depth2, fname, exportDataFlag)